Human population and statistical genetics

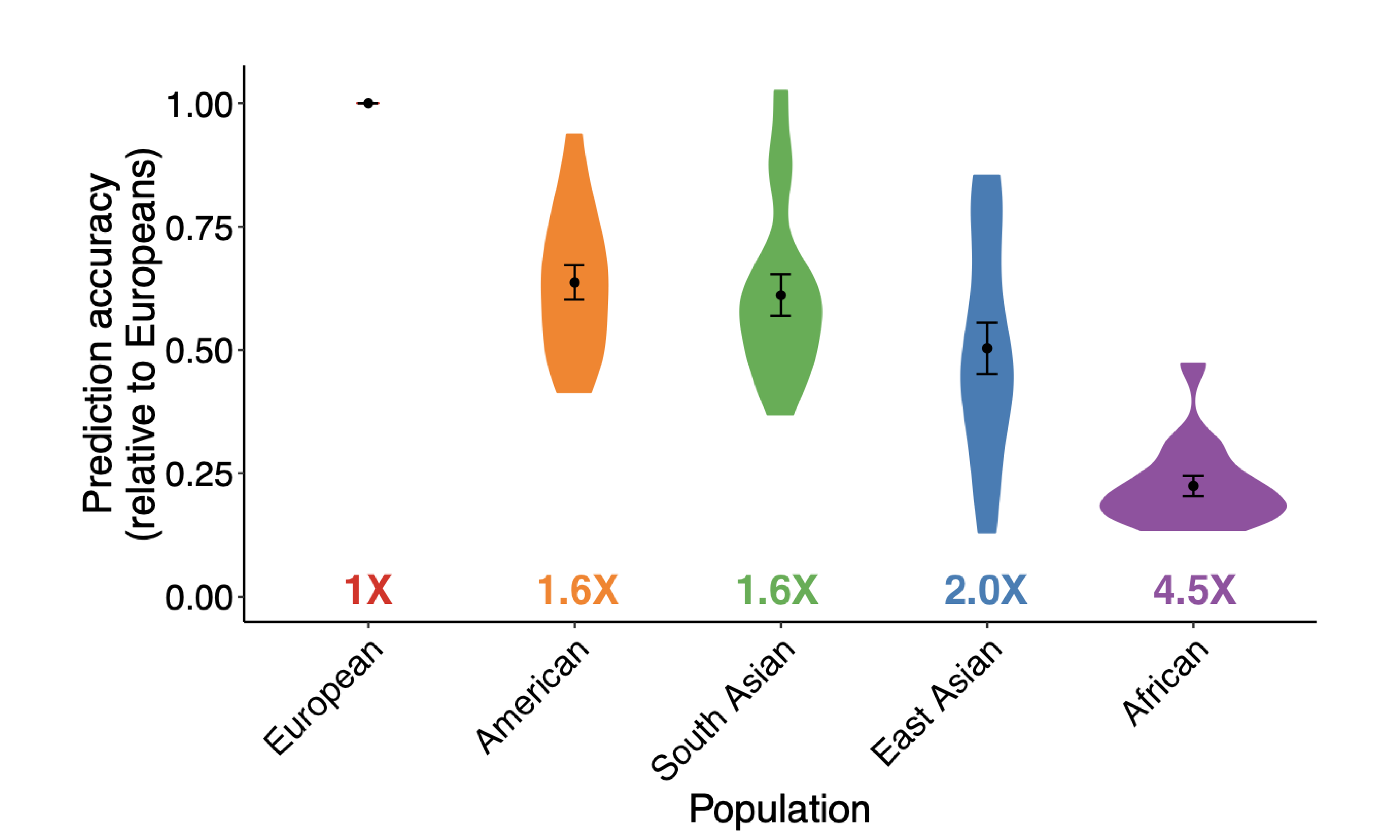
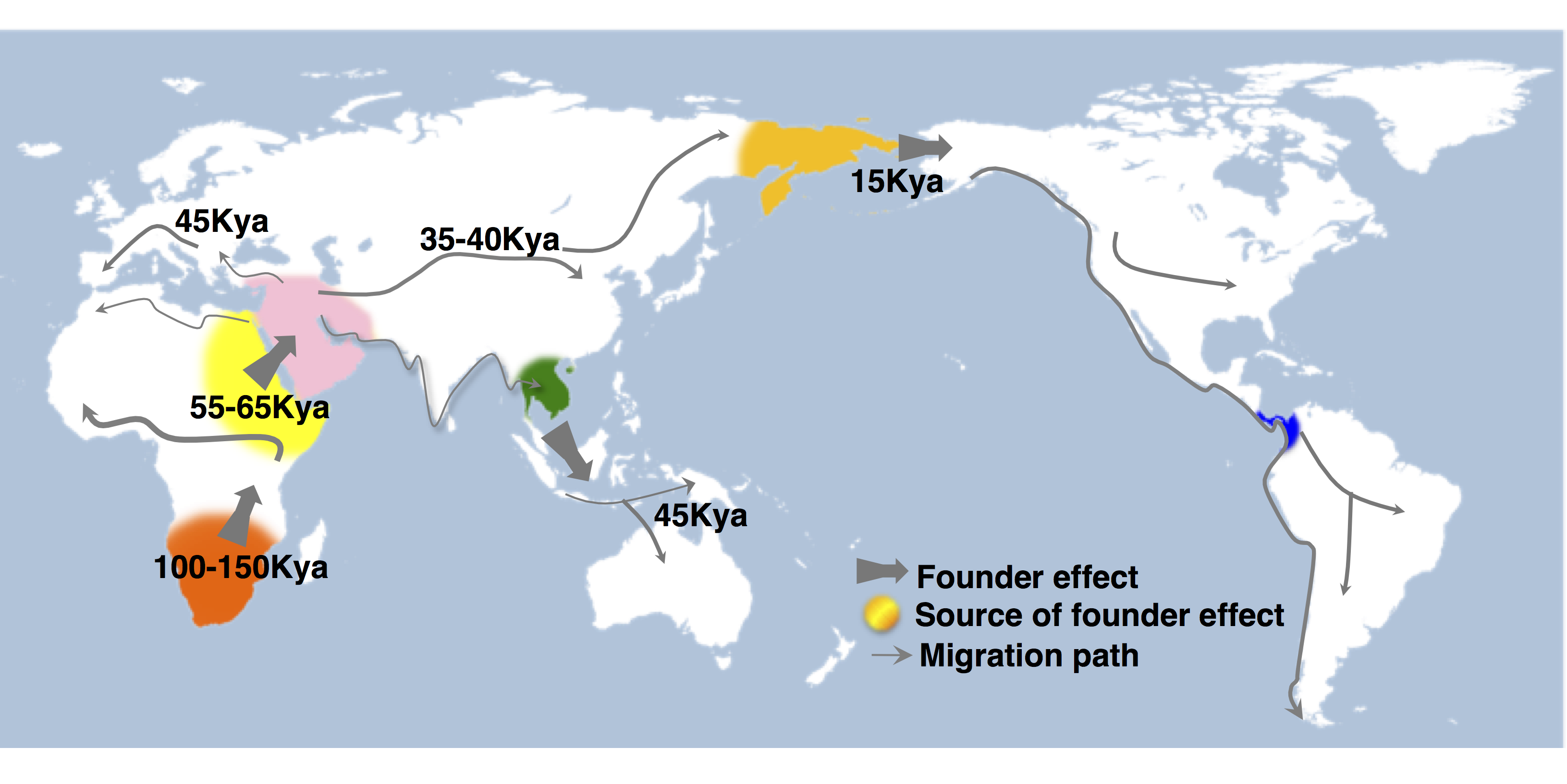
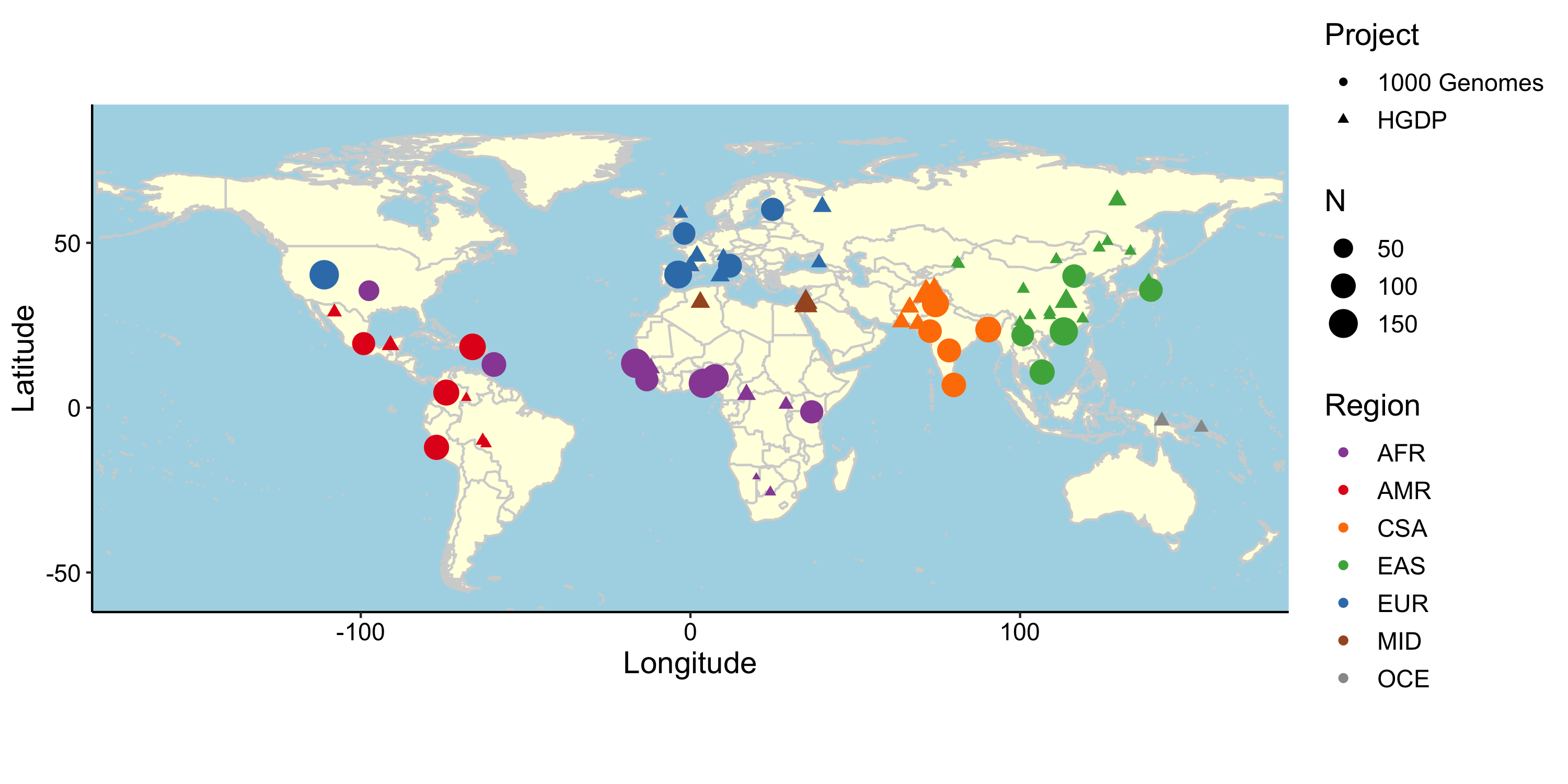
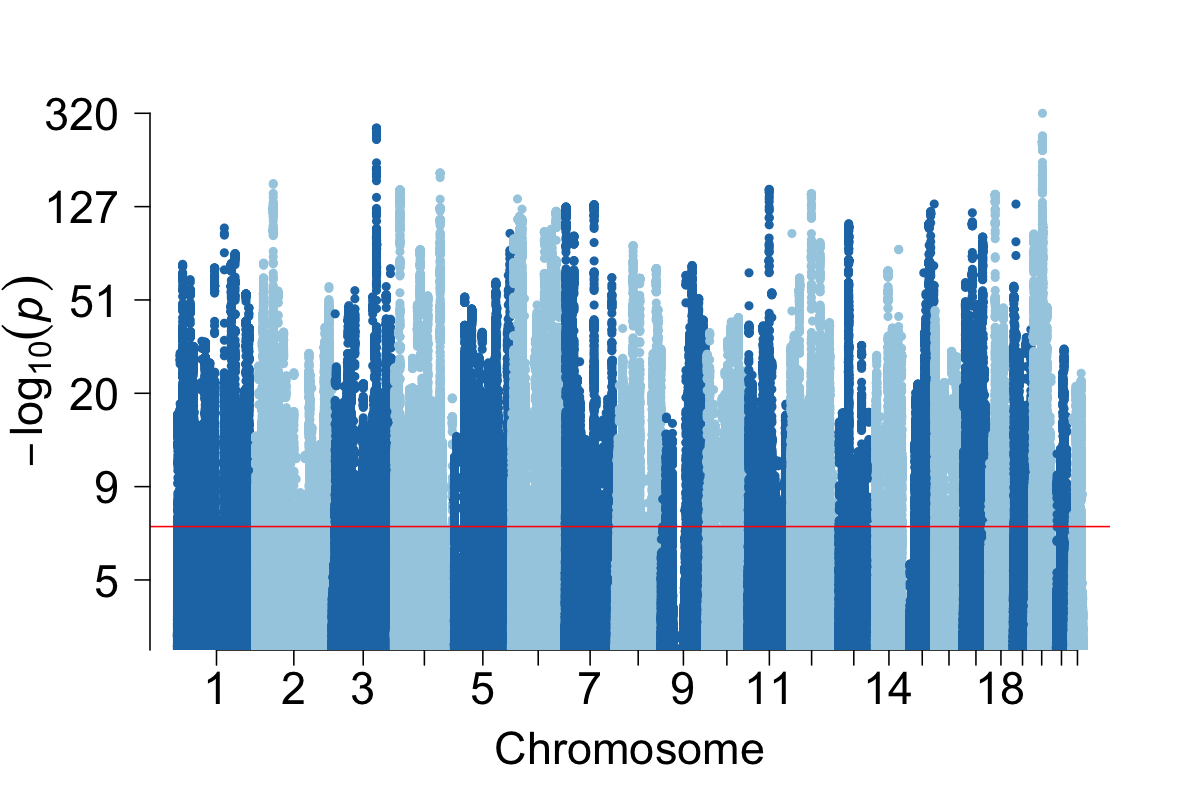
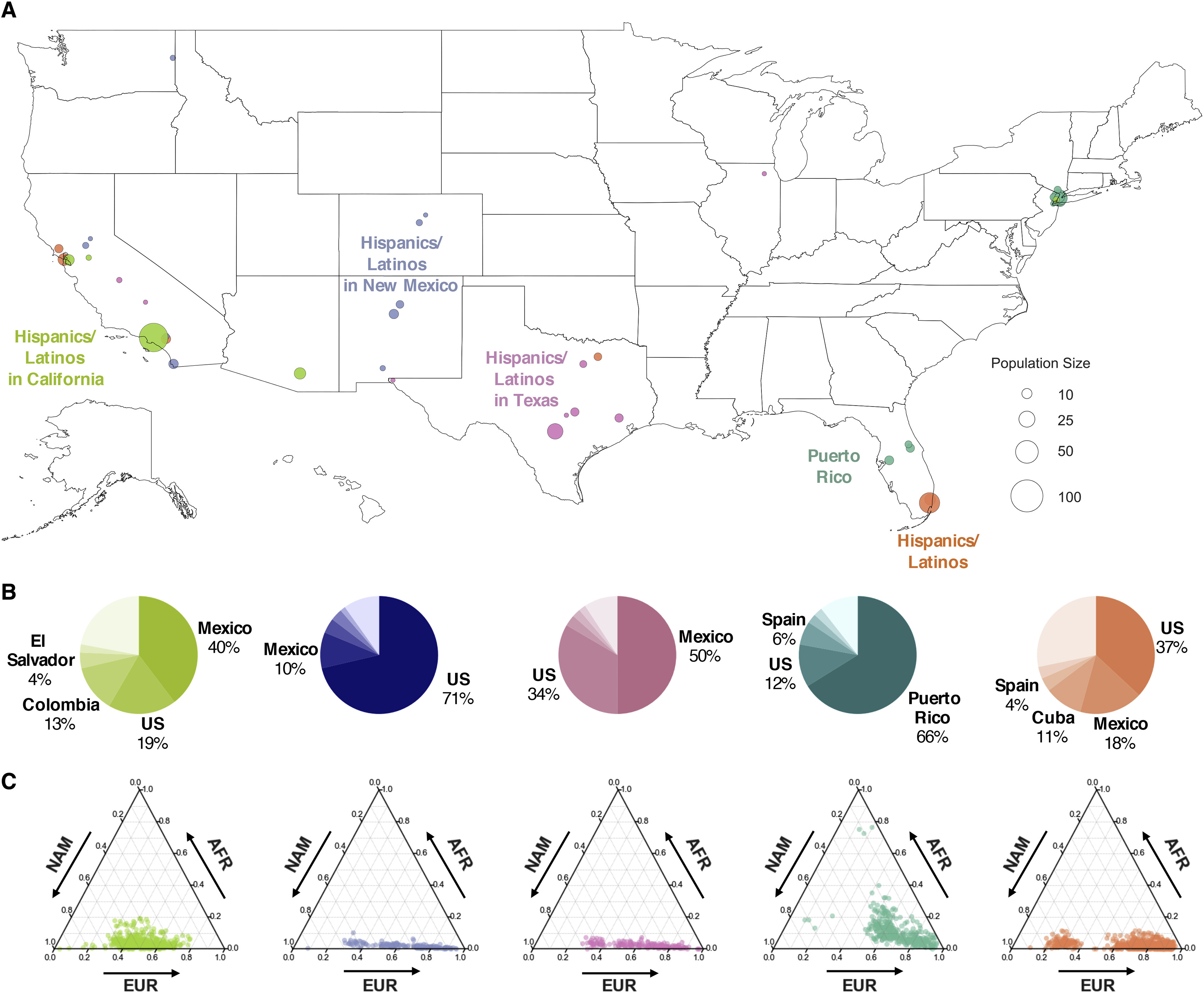
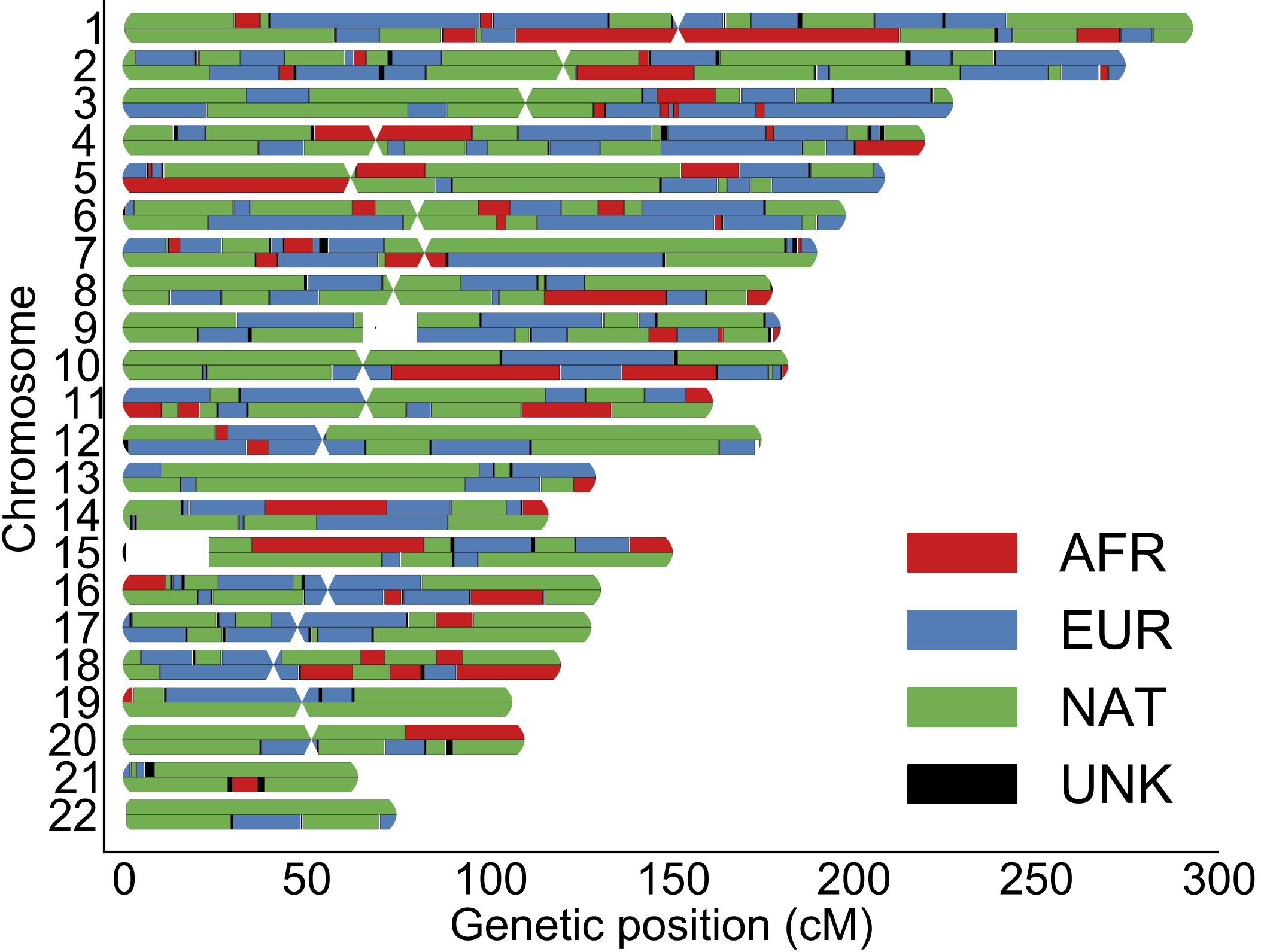
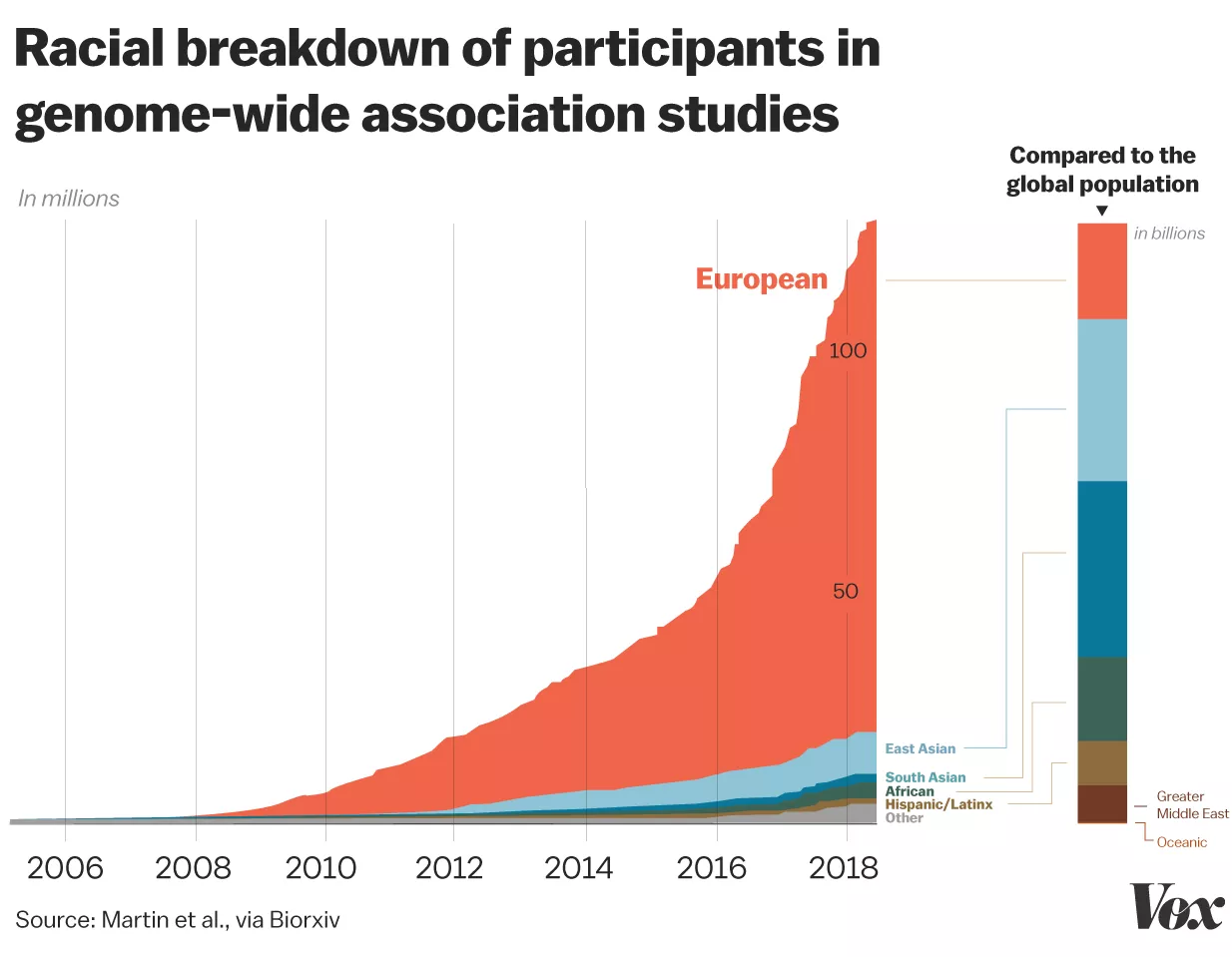
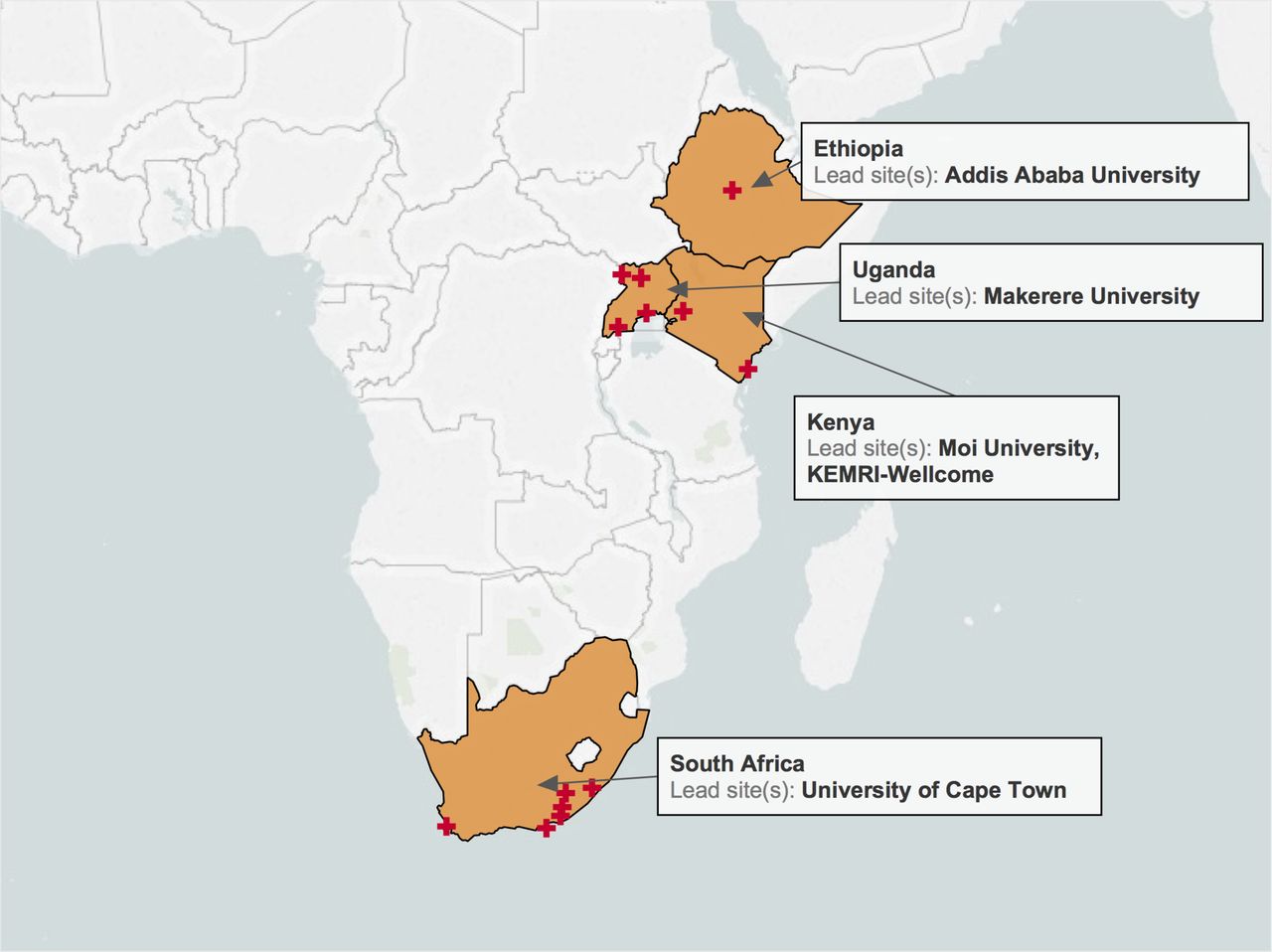
We investigate how human history shapes global genetic and phenotypic diversity, and the role these forces play in predisposing people to various complex disease risks and evolutionarily important traits. We are also particularly interested in conducting increasingly diverse studies to reduce health disparities induced by vast Eurocentric genetic study biases. We are based in the Analytic & Translational Genetics Unit in Massachusetts General Hospital as well as the Stanley Center for Psychiatric Research at the Broad Institute.
Research Areas